Exploring the Molecular Basis: Insights into Cellular Functionality and Regulation
Genetic material is the substance which not only controls the formation and expression of traits in an organism but can also replicate and pass on from a cell to its daughter cell or from one generation to next. The major steps in DNA fingerprinting
Nucleic acids are of two types –
DNA (deoxyribonucleic acid) and RNA (ribonucleic acid).
In all organisms, DNA serves as the carrier of genetic information which expresses itself through RNA but in viruses either DNA or RNA can serve as genetic material.
DNA AS GENETIC MATERIAL
The most conclusive evidence in support of DNA as the genetic material came from the following three avenues of approach on microorganisms – the transformation of bacteria, mode of infection of bacteriophages and conjugation of bacteria.
Frederick Griffith conducted a series of experiments with bacteria Streptococcus pneumoniae. He experimented with smooth (S) or virulent and rough (R) or nonvirulent strains of Streptococcus pneumoniae.

In the last experiment, Griffith did not inject living S-type into mice but living S-type appeared in the blood and caused the death of mice. Griffith concluded that there was some factor in heat-killed S-type that transformed live R-type into live S-type. This must be due to the transfer of the genetic material. However, the biochemical nature of genetic material was not defined from his experiments.
In 1944, Avery, McCarty and MacLeod fractionated the killed S-type bacteria into three components-DNA, carbohydrate and protein.
DNA fraction was further divided into two parts: one with deoxyribonuclease or DNase and the other without it. The four components were then added to separate culture tubes containing R-type bacteria as shown below:

They also discovered that protein-digesting enzymes (proteases) and RNA-digesting enzymes (RNases) did not affect transformation, so the transforming substance was not a protein or RNA. Digestion with DNase did inhibit transformation, suggesting that the DNA caused the transformation. They concluded that DNA is the hereditary material.
The unequivocal proof that DNA is the genetic material came from the experiments of Alfred Hershey and Martha Chase (1952). They worked with viruses that infect bacteria called bacteriophages. Hershey and Chase worked to discover whether it was protein or DNA from the viruses that entered the bacteria.
They incorporated radioactive isotope of phosphorus (32P) into phage DNA and that of sulfur (35S) into proteins of a separate phage culture. These phage types were used independently to infect the bacterium Escherichia coli. After some time, this mixture was agitated on a blender to separate the empty phage capsids from the surface of bacterial.
When 35S was used, all radioactive material was limited to empty viral protein coats.
Bacteria that were infected with viruses that had radioactive DNA were radioactive indicating that DNA was the material that passed from the virus to the bacteria.

These results indicated that the DNA of the bacteriophage and not the protein enters the host, where viral replication takes place. Therefore, DNA is the genetic material of bacteriophage.
DNA
DNA is the largest macromolecule which is composed of small monomeric units called nucleotides. The length of DNA is defined as a number of nucleotides present in it which is the characteristic of an organism.
Each nucleotide is made up of a pentose sugar (deoxyribose type), a phosphate group and a nitrogenous base.
A subunit composed of only sugar and the nitrogen base is known as a nucleoside.
The nitrogenous base is linked to pentose sugar through N-glycosidic linkage to form a nucleoside.
The four nucleosides differ from each other in the type of the base, which could be adenine (A), guanine (G), thymine (T), or cytosine (C).
The adenine and guanine are the purines and thymine and cytosine are the pyrimidines. (In RNA, uracil is present instead of thymine).
In a nucleotide, N-base is attached to deoxyribose sugar at C-1 and phosphoric acid is attached at C-5 of sugar.
Adjacent nucleotides are joined together by phosphodiester bonds between C–3 and C–5 of different deoxyribose sugars of two adjacent nucleotides (3′-5′ phosphodiester linkage) to form a polynucleotide chain.
The polynucleotide chains show polarity as one end of the chain has a sugar residue with C-3 not linked to another nucleotide having a free 3′-OH group and the other end has sugar residue with C-5 linked to a phosphate group (not linked to another nucleotide). These are named as 3’ and 5’ ends (three and five prime ends) of polynucleotide chain, respectively.
The two polynucleotide chains are antiparallel to each other and are held together by hydrogen bonding between specific pairs of purines and pyrimidines.
The pairing is always between A and T, and G and C. There are two hydrogen bonds between A and T and three between G and C.
The stacking of bases creates two types of grooves called major and minor grooves.
Both polynucleotide strands in a double helix remain separated by a distance of 20Å. The coiling of double helix is right-handed and one complete turn occurs every 34Å so the pitch of one turn is 34Å.
Distance between base pairs is 3.4Å and since the pitch of one turn is 34Å, so there are 10 base pairs in each turn.
Chargaff’s rules about DNA
In 1950, Erwin Chargaff formulated important generalizations about DNA structure, these generalizations are called Chargaff’s rules in his honor. They are summarised as follows:
– The purines and pyrimidines are always in equal amounts, i.e., A + G = T + C.
– The amount of adenine is always equal to that of thymine, and the amount of guanine is always equal to that of cytosine, i.e., A = T and G = C.
– The base ratio A + T / G + C may vary from one species to another, but is constant for a given species.
DNA packing in prokaryotes is carried out with the help of RNAs and nonhistone basic proteins like polyamines.
DNA packing in eukaryotes is carried out with the help of lysine and arginine-rich basic proteins called histones. The unit of compaction is the nucleosome. Histones are of five types-H1, H2A, H2B, H3 and H4. Four of them (H2A, H2B, H3 and H4) are organized to form a unit of eight molecules called histone octamer. The negatively charged DNA is wrapped around the positively charged histone octamer to form a structure called a nucleosome.
A typical nucleosome contains 200 bp of DNA helix. Nucleosomes constitute the repeating unit of a structure in nucleus called chromatin, (thread-like stained bodies seen in nucleus). The nucleosomes in chromatin are seen as ‘beads-on-string’ structures when viewed under an electron microscope.
In a typical nucleus, some regions of chromatin are loosely packed (and stain light) and are referred to as euchromatin, which is transcriptionally active chromatin. The chromatin that is more densely packed and stains dark is called as heterochromatin, which is inactive.
RNA
Ribonucleic acid (RNA) is a single-stranded structure consisting of an unbranched polynucleotide chain, but it is often folded back on itself forming helices. The four nitrogenous bases found in RNA are adenine, cytosine, guanine and uracil. Uracil is present in RNA at place of thymine. In RNA, every nucleotide residue has an –OH group present at 2′ position in the ribose.
Types of RNA
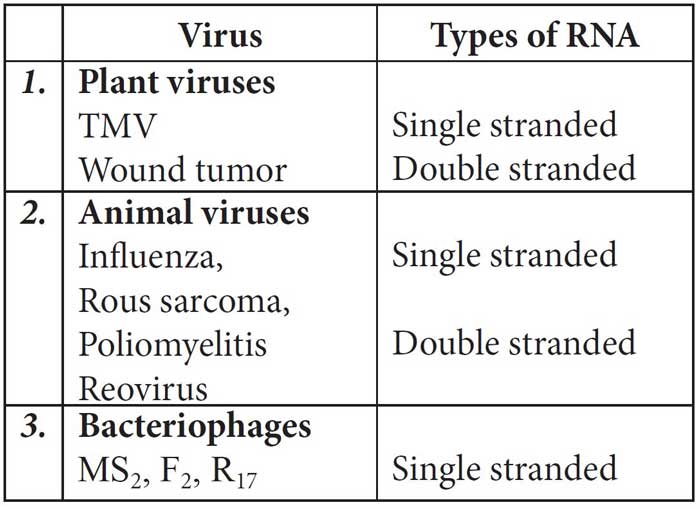
The RNA may be mainly of two types – genetic RNA and non-genetic RNA. Genetic RNA is seen in most of the plant viruses and some animal viruses, e.g., in TMV, poliovirus, influenza virus, etc., RNA acts as genetic material.
Table: Different RNA viruses and the nature of genetic RNA associated with them
On the basis of molecular size and function, three main forms of nongenetic RNA are –mRNA, tRNA and rRNA.
mRNA constitutes about 2-5% of cellular RNA; tRNA is about 15% and rRNA about 70-80%.
Messenger ribonucleic acid (mRNA) or informational RNA is a molecule of RNA that is transcribed from a gene and then translated by ribosomes in order to manufacture protein.
tRNA or transfer RNA is also known as soluble RNA or adaptor RNA. tRNAs are smallest, bearing 70-80 nucleotides and sedimentation Coefficient of 4S.
Ribosomal RNA is a component of the ribosomes, the protein synthetic factories in the cell. It is formed in nucleolus. rRNA is the most stable type of RNA.
DNA v/s RNA
DNA is a better genetic material than RNA because of the following reasons:
DNA is chemically less reactive and structurally more stable as its nucleotides are not exposed except when they have to express their effect.
– Presence of thymine in DNA instead of uracil, provides stability to DNA.
– Hydrogen bonding between purines and pyrimidines and their stacking make DNA more stable for storage of genetic information.
– DNA is capable of undergoing slow mutations required of genetic material.
– It possesses the power of repairing.
Since DNA is more stable while RNA is more reactive, both the types of nucleic acids have been retained in genetic expression.
DNA Replication
DNA replication is the unique process of making an identical copy of a double-stranded DNA, using existing DNA as a template for the synthesis of new DNA strands prior to cell division (in S phase of cell cycle).
Meselson and Stahl (1958) proved that DNA replicates by semi-conservative method by experimenting on E.coli. They grew E.coli for many generations in medium containing the heavy isotopes of nitrogen, i.e., 15N. They grew bacterial cells having DNA labelled with 15N in 14N medium and found that F1 generation has DNA density intermediate between the two.
This implied that, the newly synthesised DNA possess one strand contributed by parent DNA and other newly synthesised.
Replication of DNA is energetically highly expensive. It requires a set of enzymes, mainly DNA polymerases. These enzymes catalyse polymerisation of a large number of nucleotides very accurately in a very short time. Replication requires abundant energy that comes from breakdown of triphosphates of deoxyribonucleotides. Enzyme helicase acts over the Ori site and unwinds the two strands of DNA. The separated strands are stabilised by means of single stranded binding proteins (SSBPs). Unwinding creates tension in the uncoiled part by forming more supercoils. Tension is released by enzymes topoisomerases.
They cause nicking of one strand of DNA (for removing coils) and resealing the same. Whole of DNA does not open in one stretch due to very high energy requirement but the point of separation proceeds slowly towards both the directions. It gives the appearance of Y-shaped structure called replication fork.
A small strand of RNA, called RNA primer, is synthesised at the 5′ end of new DNA strand.
It is essential for initiation of new DNA chains as it is required by DNA polymerases to add nucleotides. DNA polymerases catalyse polymerisation only in one direction that is 5′ _ 3′. Consequently, on one strand called leading strand (the template with polarity 3′ _ 5′), the replication is continuous, while on the other strand called lagging strand (the template with polarity 5′ _ 3′), it is discontinuous.
Discontinuous replication occurs because only a small stretch of template exposes at a time.
The discontinuously synthesised fragments are later joined by the enzyme DNA ligase.
Central Dogma
Crick (1958) proposed the central dogma of molecular biology. Central dogma is the unidirectional flow of information from DNA to RNA and from RNA to polypeptide.
H. Temin and Baltimore (1970) introduced the concept of reverse central dogma, i.e., formation of DNA from RNA. It is also called teminism and occurs in retroviruses.
Transcription
The process of copying genetic information from one strand of the DNA into RNA is termed as transcription.
The segment of DNA that takes part in transcription is called transcription unit.
It has three components a promoter, the structural genes and a terminator. Besides a promoter, eukaryotes also require an enhancer.
Promoter is located upstream of structural gene. Terminator region is present downstream (3′ end of coding strand) of structural gene. The promoter has an AT rich region, called TATA box also called Pribnow box.
In structural gene transcription can occur only in 5′→ 3′ direction. The strand of DNA taking part in transcription is called the template strand.
In prokaryotes RNA polymerase is only of one type and can transcribe all types of RNAs. It has sigma subunit attached with core enzyme having 5 subunits – b′, b, a1, a2 and w.
In eukaryotes, three major classes of RNA polymerases are found in the nucleus. RNA polymerase I synthesises precursors for the large ribosomal RNAs. RNA polymerase II synthesises the precursors for mRNAs and small nuclear RNAs. RNA polymerase III participates in the formation of tRNAs and 5S rRNAs.
The DNA segment which codes for polypeptide chain is called cistron. The structural gene in a transcription unit could be monocistronic (mostly in eukaryotes) which codes for only one protein or polycistronic (mostly in bacteria or prokaryotes) which codes for many proteins.
Prior to transcription the nucleotides are activated through phosphorylation. Enzyme phosphorylase is required along with energy for this.
Transcription takes place in three steps – initiation, elongation and termination.
The first step in transcription is the binding of RNA polymerase to DNA. The specific region on the DNA where the enzyme binds to form closed promoter complex is known as the promoter region. It is located on the 5′ end of the gene to be transcribed which signals where to start RNA synthesis. It also determines which DNA strand is to be transcribed. Thus a promoter region has RNA polymerase recognition site and RNA polymerase binding site.
When the closed promoter complex forms, DNA remains double helical. It then isomerizes and causes unwinding and separation of DNA strands to form open promoter complex, a highly stable structure.
RNA polymerase contains two nucleotide-binding sites: initiation site and elongation site.
– Initiation site binds only with purine triphosphates (ATP and GTP). The initiating nucleoside triphosphate binds to the enzyme in open promoter complex and forms hydrogen bond with a complementary DNA base (base pairing).
– Elongation site is then occupied by a nucleoside triphosphate that is selected strictly on the basis of its ability to form hydrogen bond with the next base in DNA strand. The two nucleotides are then joined together and the first base pair is released from initiation site.
In prokaryotes, the transcription product directly function as mRNA but in eukaryotes the result of transcription is hnRNA.
In eukaryotes, hnRNA contains both the exons (expressing sequences) and the introns (interrupting sequences) that are nonfunctional. Hence, it is subjected to a process called splicing where the introns are removed and exons are joined in a defined order.
hnRNA undergoes additional processing called as capping and tailing. In capping, methyl guanosine triphosphate is added to the 5′ – end of hnRNA. In tailing, adenylate residues are added at 3′ – end in a template-independent manner.
Genetic Code
Sequence of nitrogenous bases or nucleotides in a polynucleotide chain (DNA molecule) which determines sequence of amino acids in a polypeptide chain is called genetic code. The 64 distinct triplets determine the 20 amino acids. The DNA sequence of a gene is divided into a series of units of three bases called codon.
The salient features of genetic code are as follows:
– The codon is triplet. 61 codons code for amino acids and 3 codons (UAA, UAG and UGA) do not code for any amino acids, hence they function as stop codons.
– One codon codes for only one amino acid, hence, it is unambiguous and specific.
– Some amino acids are coded by more than one codon, hence the code is degenerate.
– The codon is read in mRNA in a continuous fashion. There are no punctuation.
– The code is nearly universal: i.e., found in all living organisms.
– AUG has dual functions. It codes for methionine (met), and it also act as initiation codon.
Wobble hypothesis was given by F.H.C. Crick (1966). According to this, the third nitrogenous base of a codon is not much significant and codon is specified by the first two bases. Hence the same tRNA can recognize more than one codons differing only in third position.
TRANSLATION
Translation or protein synthesis is a process during which the genetic information stored in the sequence of nucleotides in an mRNA molecule is translated, following the dictation of the genetic code, into the sequence of amino acids in the polypeptide which requires the functions of a large number of macromolecules.
Protein synthesis occurs over the ribosomes (protein factories). Each ribosome has two unequal parts, small and large. The larger subunit of ribosomes has a groove for pushing out the newly formed polypeptide and protecting the same from cellular enzymes.
The smaller subunit fits over the larger one like a cap but leaves a tunnel for mRNA. The two subunits come together only at the time of protein formation. The phenomenon is called association. Mg2+ is essential for it. Ribosomes usually form groups during active protein synthesis, known as polyribosomes or polysomes.
There are three reactive sites in a ribosome –P-site (peptidyl transfer or donor site.) A-site (amino-acyl or acceptor site) and E or exit site.
P-site is jointly contributed by both ribosomal subunits. A-site and E-site are largely confined to the larger subunit of ribosome.
The synthesis of polypeptide can be considered in terms of initiation, elongation and termination stages. These fundamental processes have additional stages: activation of amino acids before their incorporation into polypeptide and the post-translational processing of the completed polypeptide. Both these processes paly important roles in ensuring the proper function of the protein product.
Amino acids are activated in the presence of ATP and linked to their cognate tRNA-a process commonly called as charging of tRNA or aminoacylation of tRNA to be more specific. The translational of mRNA begins with the formation of the initiation complex. Initiation factors are designated as IFs in prokaryotes and eIFs in eukaryotes. Elongation requires the initiation complex, aminoacyl-tRNAs, elongation factors and GTP.
Human Genome Project
With the establishment of genetic engineering techniques where it was possible to isolate and clone any piece of DNA and availability of simple and fast techniques for determining DNA sequences, a very ambitious project of sequencing human genome was launched in the year 1990 called as Human Genome Project.
Some of the important goals of HGP were as follows:
– Identify all the genes present in the human genome.
– Determine the sequences and numbers of all the base pairs that make up the human genome.
– Store this information in databases.
– Identify various genes that cause genetic disorders.
– Improve tools for data analysis.
– Transfer related technologies to other sectors, such as industries.
– Address the ethical, legal, and social issues (ELSI) that may arise from the project.
The human genome project was coordinated by the U.S. Department of Energy and the National Institute of Health. Additional contributions came from Japan, France, Germany, China and others.
The basic strategies of human genome project are mapping, sequencing and functional analysis.
Mapping is to prepare genetic and physical maps of the human genome.
Sequencing the entire genomic DNA of humans is limited because of the limitations of the sequencing techniques.
Functional analysis is to relate each gene with its function.
DNA Fingerprinting
DNA fingerprinting involves identifying differences in some specific regions in DNA sequence called as repetitive DNA because in these sequences, a small stretch of DNA is repeated many times.
These repetitive DNA are separated from bulk genomic DNA as different peaks during density gradient centrifugation. The bulk DNA forms a major peak and the other small peaks are referred to as satellite DNA. Depending on the base composition (A: T rich or G:C rich), length of the segment, and a number of repetitive units, the satellite DNA is classified into many categories, such as micro-satellites, mini-satellites etc.
These sequences normally do not code for any proteins and show a high degree of polymorphism hence form the basis of DNA fingerprinting.
Minisatellites or Variable Number Tandem Repeat (VNTRs) are very specific in each individual and vary in number from person to person but are inherited. Each individual inherits these repeats from his/her parents which are used as genetic markers in a personal identity test.
The major steps in DNA fingerprinting are as follows:
– DNA is extracted from the cells. DNA can be amplified by PCR or polymerase chain reaction.
– DNA is cut into fragments with restriction enzymes.
– Chopped DNA fragments are passed through electrophoresis. The separated fragments can be visualised by staining them with a dye.
– Double-stranded DNA is then split into single-stranded DNA using alkaline chemicals.
– Separated DNA sequences are transferred from gel onto a nitrocellulose or nylon membrane.
– The nylon sheet is then immersed in a bath, where probes or markers radioactive synthetic DNA segments of known sequences are added. _e probe hybridises VNTR (Southern blotting).
– X-ray film is exposed to the nylon sheet gives dark bands at the probe sites. Thus hybridised fragments are detected by autoradiography.
– DNA fingerprinting helps in solving parentage dispute as well as identifying criminals.
